click image to see full view
Miscellaneous
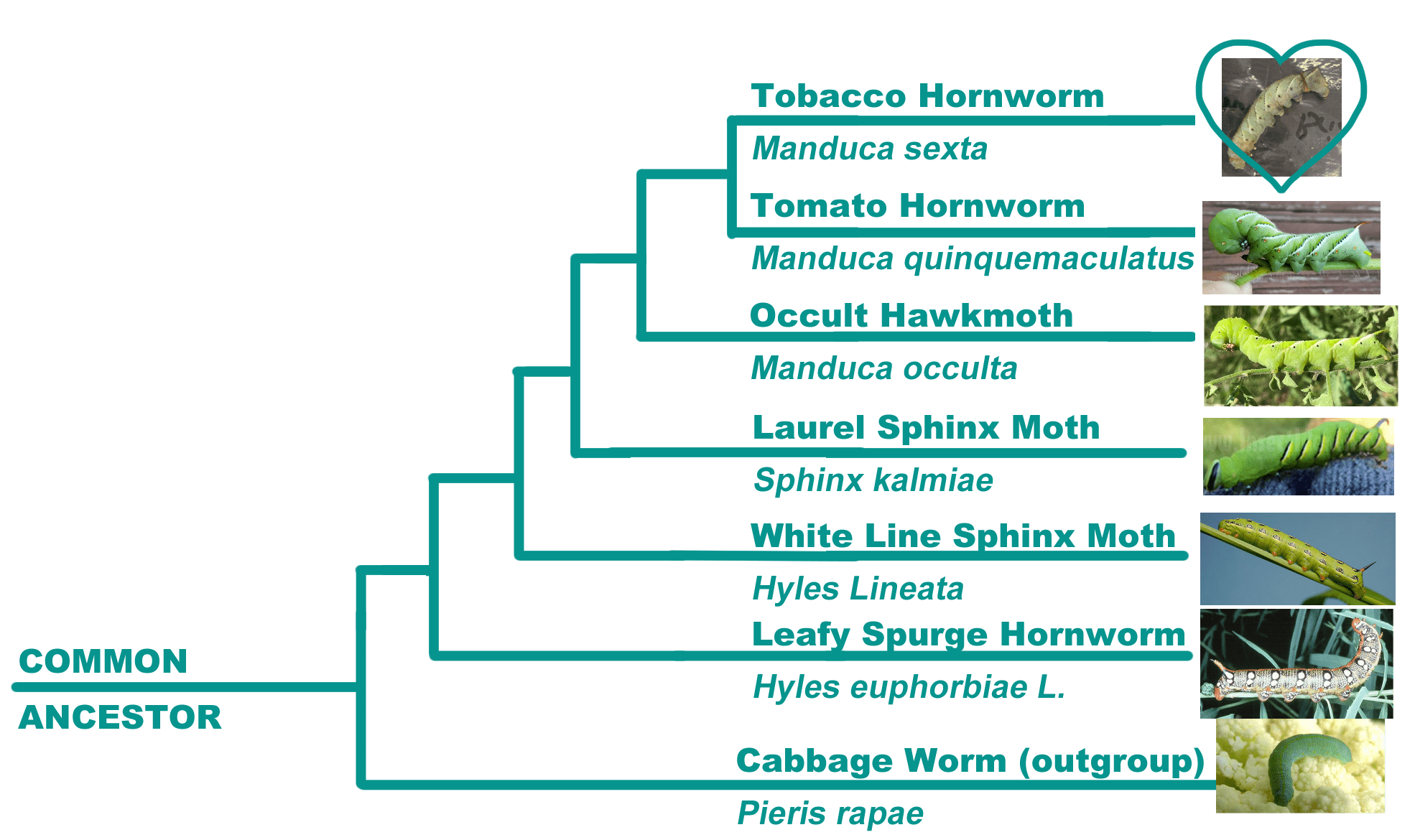
In one of my college biology classes we had to create a phylogenetic tree based on a DNA sequence that we got from our own specimen. I provided a hornworm from my sibling's reptile's meal box, which we then extracted some mitochondrial DNA from (mitochondrial COI, for those in the know). The extracted DNA was sent to a lab to be sequenced. We then used a DNA database to compare the sequence we got back from the lab to find out which species our sequence was most closely related to. We had incorrectly assumed our specimen was a tomato hornworm at first (Manduca quinquemaculatus), but both the DNA and the company itself after I emailed it confirmed that it was actually a tobacco hornworm (Manduca sexta) which are incredibly closely related. We then compared these two specimens to other hornworms, then used a cabbage worm as our reference outgroup. We used DNA Subway and their database to construct the formal phylogenetic tree. While making the poster I was charged with the graphic design and thus made this phylogenetic tree of our chosen species. We were one of the few groups that actually got DNA to extract, so that was pretty cool. Our sequence only had one nucleotide difference from the standard tomato hornworm, but twenty differences from a tobacco hornworm. The number of differences increases as you move outward. I forgot exactly how many differences there were between our specimen and the cabbage worm, but it probably was well over a hundred. We used mitochondrial COI for this sequence because every animal in the world happens to have this particular DNA in them in some form, so it's a good reference for comparing the evolutionary lines of animal species. For a plant you used the Rubisco gene, which codes chlorophyll. The functional part of the COI gene remains the same throughout different animal species, but because of gradual changes in DNA over time through evolution the buffer parts of the gene that are in place to protect the functional ones change quite a bit. The theory is that if a specimen has fewer differences in its buffer sequence, then it is more closely related to the comparison species. It was a fun project! I would link the poster for the project, but it has my name and college on it and I'd rather not be doxxed. I'm also not a genetic biologist. This explanation of the project and its working ideas is heavily simplified and based on what I learned about and from it, and I also did not explain the entire process. DNA barcoding is fascinating, though. I recommend looking into it if you have time.
DNA Subway A site about hornworms More about DNA Barcoding